-
Project IDT2021-152
-
RFP Year2021
-
Awarded Amount$838,587DiseaseMalariaInterventionDrugDevelopment StageTarget IdentificationCollaboration PartnersFIMECS, Inc. , National Center for Genetic Engineering and Biotechnology (BIOTEC)
Introduction and Background of the Project
1. Introduction
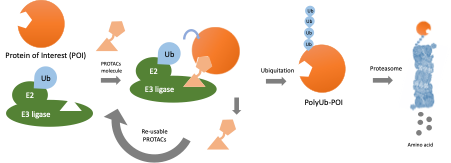
Malaria is one of the most widespread infectious diseases in which over 200 million infections occur annually worldwide. Current antimalarial drugs do not work well against malarial parasite. Antimalarial drugs act by inhibiting a biochemical process in the parasite, for example, an enzyme target, whereas the drug must be specific to the target to prevent toxic side effects. This specificity requirement is an Achilles’ heel as drug-resistant parasites with small genetic changes that prevent the drug from binding. One solution to drug resistance is to develop new drugs against new targets. However, developing new drugs is challenging because there are few suitable drug targets. A new drug design approach is needed to expand the repertoire of antimalarial targets and the drug arsenal to exploit previously untouched vulnerabilities in the parasite. Instead of using drugs that act as inhibitors of target functions, drugs designed as protein degrader, also known as PROteolysis-TArgeting Chimeras or PROTACs, can destroy target proteins. The protein degrader work by hijacking the proteasome (a protein complex present in every cell that naturally breaks down old or damaged proteins) to degrade a target protein. Protein degraders are designed with one “warhead” that binds the target and another that binds ubiquitin E3 ligase, a protein that marks other proteins for degradation. This approach has the advantage that the protein degraders can bind anywhere on the target, without the restriction of binding to sites important for target function. Therefore, the protein degraders can be designed against proteins previously not considered as drug targets. PROTAC antimalarials acting on new targets throughout the parasite life cycle could be important tools for the eradication of malaria. However, no small molecule PROTAC antimalarial currently exists because we lack knowledge of malaria parasite ubiquitin E3 ligases, and no chemical “warhead” is available that can recruit a parasite ubiquitin E3 ligase for hijacking the parasite’s proteasome.
2. Project objective
To identify a chemical warhead(s) that can recruit a parasite ubiquitin E3 ligase(s) to degrade a target parasite protein, which will constitute a platform for the design of protein degrader antimalarials.
3. Project design
We will design and synthesize a library of protein degraders for degradation experiments. The test compounds will be designed with various chemical warheads against a variety of ubiquitin E3 ligases joined to a warhead specific to the Plasmodium parasite bifunctional dihydrofolate reductase-thymidylate synthase, a well-studied parasite protein. The protein degrader that trigger target protein degradation will be used for designing follow-on compounds, including probe compounds for biochemical characterization of Plasmodium ubiquitin E3 ligase(s) that interact with the ubiquitin E3 ligase warhead and those protein degraders for optimizing the degradation of target.
How can your partnership (project) address global health challenges?
There is a growing concern that malaria will become resurgent owing to the evolution and spread of drug-resistant malaria parasites. New drugs with novel mechanisms of action are needed to counter this threat. These new drugs must act against all stages of the parasite life cycle, including stages not targeted by current drugs. Protein degrader antimalarial drugs could fulfill these needs by making many new targets accessible to chemotherapy. If our approach can be demonstrated for antimalarial drugs, the same approach could be applied for other neglected parasitic diseases, such as Leishmaniasis.
What sort of innovation are you bringing in your project?
If successful, we will establish a platform for protein degraders as antimalarial drugs that act by degrading parasite protein targets. This would represent the first small molecule protein degradation platform for drugs against parasitic diseases that target non-human proteins.
Role and Responsibility of Each Partner
The FIMECS company (Japan) will synthesize a library of protein degraders for initial screening. The BIOTEC Institute (Thailand) will perform protein degradation experiments and the protein degrader optimization of follow-on compounds.
Others (including references if necessary)
Final Report
1. Project objectives
PROteolysis-TArgeting Chimeras (PROTACs) are drugs that mediate degradation of target proteins by hijacking the ubiquitin proteasome system (UPS). Unlike conventional drugs, PROTACs do not require high-affinity binding to mediate target degradation, which is advantageous as they can be directed against more targets and are less prone to resistance. However, no PROTACs are available against infectious diseases such as malaria. The objective of the project was to identify small molecules capable of recruiting the Plasmodium falciparum malaria parasite UPS for target degradation towards developing PROTAC antimalarials.
2. Project designHeterobifunctional compounds were synthesized comprising a ligand binding to the P. falciparum target dihydrofolate reductase-thymidylate synthase joined to various UPS-recruiting ligands. The compounds were assessed for their ability to induce target degradation in P. falciparum by western blotting. From the frontrunner compounds with target degradation activity, photoactivatable probe derivatives were synthesized for biochemical enrichment and identification of the UPS-recruited proteins. Frontrunner compounds were used in co-immunoprecipitation experiments in an alternative approach for identifying UPS-recruited proteins. Finally, we synthesized homo-bivalent compounds comprising two UPS-recruiting ligands (present in frontrunner heterobifunctional compounds) joined by different linkers and tested them for antimalarial activity.
3. Results, lessons learned
The success of the project hinged on the collaborative efforts of partners with complementary resources and expertise. FIMECS Inc., the Japanese partner, leveraged their specialized resources in PROTAC discovery. They designed and synthesized custom heterobifunctional compounds and conducted testing of the drug metabolism and pharmacokinetic (DMPK) properties for these compounds. The non-Japanese partner, BIOTEC, drew upon their experience in conventional target-based antimalarial drug research for designing, synthesizing, and testing compounds for their biological activities. Despite the challenges posed by limited knowledge regarding UPS-recruitment in P. falciparum, we identified compounds with distinct UPS-recruiting ligands that induced rapid target degradation (more than 50% reduction in 4 h) and exhibited potent antimalarial activity (50% growth inhibition concentration less than one micromolar). To progress these compounds into potential drugs, it is imperative to understand their mode of action, particularly in terms of the recruited UPS component(s). Initial attempts to elucidate the mode of action using photoactivable derivatives proved unsuccessful due to nonspecific protein interaction. However, employing the alternative co-immunoprecipitation approach enabled us to identify the NOT4 E3 ligase as a potential recruited UPS factor. Homo-bivalent compounds possessed antimalarial activity comparable to frontrunner heterobifunctional compounds. This suggests that the NOT4 protein itself could serve as a target for PROTACs in antimalarial drug development. Furthermore, the homo-bivalent compounds exhibited favorable DMPK properties characterized by high solubility and metabolic stability. The promising results from the project underscore the value of further investment in the development of PROTAC-based antimalarials. Future work includes validating the mechanism of action and gaining a comprehensive understanding of how bifunctional molecules engage the P. falciparum UPS for target degradation.
Investment
Details
Identification and Validation of potential Plasmodium E3 Ligases for PROTAC Platform




